A U-Net model's ability to successfully generate and estimate pixel-level lung thickness maps and total lung volumes from chest radiographs (CXRs) may be helpful for diagnosing and preventing the progression of pulmonary diseases during routine CXR screenings, researchers have found.
The deep-learning model's measurements, when examined, demonstrated a strong correlation with measurements derived from CT, according to the team led by Tina Dorosti and Manuel Schultheiss of Technical University of Munich in Germany. Their results were published May 28 in Radiology: Artificial Intelligence.
The study investigated pixel-level lung thickness maps and total lung volume (TLV) estimation using the U-Net trained on 7,302 synthetic x-rays, validated using 2,191 images, and applied to real radiographs.
"To the best of our knowledge, there are no deep-learning models available for pixel-level lung volume estimation of real CXRs," the team noted. The researchers listed three benefits of pixel-level thickness maps, including the following:
- Improving the accuracy of measuring of volumetric changes in pulmonary tissue, which could improve risk assessment for pulmonary resection surgery
- Proving useful in novel imaging modalities such as dark-field x-ray, where dividing the measured signal by the lung thickness improves the image contrast
- Assisting physicians in diagnosing and preventing the progression of pulmonary diseases during routine CXR screenings.
To establish the model's potential, Dorosti and colleagues simulated synthetic frontal CXRs and two-dimensional lung thickness maps from CT data and volumetric lung segmentations. The team used two publicly available datasets (the 2016 lung nodule analysis Luna16 challenge and the 2020 RSNA pulmonary embolism detection challenge). For comparability reasons, only CT scans acquired with 120 kVp were considered, they noted.
The final model was tested on 1,191 synthetic x-rays generated from the public data (Luna16, n = 131; pulmonary embolism, n = 1,060), and 72 image pairs (healthy, n = 33; chronic obstructive pulmonary disease, n = 39) obtained from the Technical University of Munich between October 2018 and December 2019.
The team assessed the U-Net model’s performance using both the synthetic and real x-rays and examined its robustness for lung nodules, pulmonary embolism, and chronic obstructive pulmonary disease in comparison to healthy lungs.
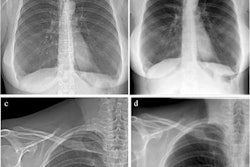
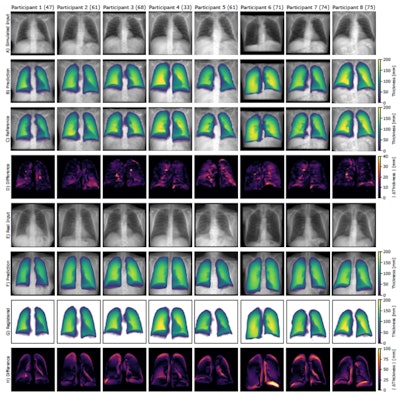 Qualitative results of eight frontal chest radiographs from the synthetic public test set, where each column represents one participant. Participants one through four are from the Luna16 test set, and participants five through eight are from the pulmonary embolism test set. (A) Simulated input radiographs, (B) predicted thickness maps, (C) reference standard thickness maps, and (D) absolute difference between the reference standard and predicted maps. The color bars for B and C indicate lung thickness in millimeters (mm). The color bar for D indicates the pixel-level estimation error in mm.Radiology: Artificial Intelligence
Qualitative results of eight frontal chest radiographs from the synthetic public test set, where each column represents one participant. Participants one through four are from the Luna16 test set, and participants five through eight are from the pulmonary embolism test set. (A) Simulated input radiographs, (B) predicted thickness maps, (C) reference standard thickness maps, and (D) absolute difference between the reference standard and predicted maps. The color bars for B and C indicate lung thickness in millimeters (mm). The color bar for D indicates the pixel-level estimation error in mm.Radiology: Artificial Intelligence
Overall, the group found that for both synthetic and real data, regardless of the pathology, their reference standard and predicted TLV results were within the previously reported CT-derived mean TLV range for healthy lungs.
The team also found the following:
- Compared with previous work on lung volume estimation with deep-learning models for real radiographs, the model achieved better results. The group reported a mean absolute error of 0.46 L, r = 0.91, compared to 0.59 L, r = 0.86.
- Overall error rates for TLV predictions were low, according to the authors. The group reported a mean squared error of 0.16 L for the synthetic public dataset, 0.20 L for the KRI dataset, and 0.35 L for the real radiograph dataset.
- TLV estimated from synthetic and real chest radiographs by the model using lung thickness maps strongly correlated with CT-derived TLV, with the team reporting r = 0.97 and 0.91, respectively; both p < 0.001.
"Since such a lung thickness map cannot be calculated from real radiographs due to the missing reference standard information, the training must be performed exclusively with simulated radiographs calculated from CT scans," Dorosti and colleagues said, emphasizing that they were still able to test the model on real, non-simulated x-rays and report promising results.
The complete study, simulation process, and all data can be found here.