Fine-tuned large language models (LLMs) show potential as proofreading applications for radiology reports, according to a study published May 20 in Radiology.
The finding is from a study in which GPT-4 and a Llama-3-70B-Instruct model demonstrated high accuracy detecting errors on a dataset of 614 chest x-ray reports with and without errors, noted lead authors Cong Sun, PhD, Kurt Teichman, of Weill Cornell Medicine in New York City, and colleagues.
“This study highlights the potential of fine-tuned large language models (LLMs) in enhancing error detection within radiology reports, providing an efficient and accurate tool for medical proofreading,” the group wrote.
Radiology reports are essential for optimal patient care, yet their accuracy can be compromised by factors like errors in speech recognition software, variability in perceptual and interpretive processes, and cognitive biases, the authors explained. Moreover, these errors can lead to incorrect diagnoses or delayed treatments, they added.
LLMs are advanced generative AI models trained on vast amounts of text to generate human language. LLMs such as GPT-4 for detecting errors in radiology reports, research on evaluating the performance of radiology-specific fine-tuned LLMs for proofreading errors remains in its early stages, the group wrote.
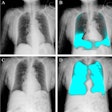
To bridge the knowledge gap, the researchers first built a training dataset and a test dataset. The training set consisted of 1,656 synthetic reports, including 828 error-free reports and 828 reports with errors. The test set comprised 614 reports, including 307 error-free reports from a large, publicly available database of chest x-rays (MIMIC-CXR), and 307 synthetic reports with errors.
Next, the researchers fine-tuned three models -- BiomedBERT, Llama-3-8Binstruct, and Llama-3-70B-Instruct – on the training set. The models were refined using zero-shot prompting, few-shot prompting, or fine-tuning strategies, the researchers noted.
Finally, using the test set, the group examined the performances of different LLMs, including the base and fine-tuned Llama-3-70B-Instruct, GPT-4–1106-Preview, and BiomedBERT models for evaluating the internal consistency of reports. All errors were categorized into four types: negation, left/right, interval change, and transcription errors.
According to the results, the fine-tuned Llama-3-70B-Instruct model achieved the best overall performance, with an overall macro F1 score of 0.780 and F1 scores of 0.769 for detecting negation errors, 0.772 for right/left errors, 0.750 for interval change errors, and 0.828 for transcription errors.
“Generative LLMs, fine-tuned on synthetic and MIMIC-CXR radiology reports, greatly enhanced error detection in radiology reports,” the group wrote.
Ultimately, the findings show that fine-tuning is crucial for enabling local deployment of LLMs while also demonstrating the importance of prompt design in optimizing performance for specific medical tasks, the researchers concluded.
In an accompanying editorial, Cristina Marrocchio, MD, and Nicola Sverzellati, MD, of the University of Parma in Italy, noted that the study represents a further step toward assessing the potential roles of LLMs in radiology, but that many questions on the wider applicability and reliability of these models remain to be addressed.
For instance, the impact of LLMs in identifying errors in radiology reports in daily clinical practice has not been evaluated, and whether they would effectively result in a lower error rate or in radiologists spending less time proofreading the report text remains to be proven, they wrote.
“Before any implementation of these systems in clinical practice is considered, a thorough evaluation of their performance and generalizability under different conditions, as well as their reliability in giving consistent answers, is essential,” Marrocchio and Nicola Sverzellati concluded.
The full study is available here.